How to Read Multiple Excel Files in R
Introduction
This mail will show you how to write and read a list of data tables to and from Excel with purrr, the functional programming package 📦 from tidyverse. In this case I will also use the packages readxl and writexl for reading and writing in Excel files, and embrace methods for both XLSX and CSV (not strictly Excel, but might also!) files.


Whilst the internet is certainly in no shortage of R tutorials on how to read and write Excel files (see this Stack Overflow thread for example), I think a purrr approach notwithstanding isn't also-known or well-documented. I find this approach to be very make clean and readable, and certainly more than "tidyverse-consequent" than other approaches which rely on lapply() or for loops. My selection of packages 📦 for reading and writing Excel files are readxl and writexl, where the advantage is that neither of them require external dependencies.
For reading and writing CSV files, I personally have switched back and forth between readr and data.table, depending on whether I have a need to practise a particular analysis in data.tabular array (see this discussion on why I sometimes use information technology in favour of dplyr). Where applicative in this post, I will point out in places where you can use alternatives from data.table for fast reading/writing.
For documentation/demonstration purposes, I'll make the package references (indicated by ::) explicit in the functions below, but it'southward advisable to remove them in "real life" to avoid code that is overly verbose.
Getting Started
The fundamental functions used in this vignette come from three packages: purrr, readxl, and writexl.
library(tidyverse) library(readxl) library(writexl) Since purrr is part of core tidyverse, we can simply run library(tidyverse). This is too convenient as we'll also utilize various functions such as group_split() from dplyr and the %>% operator from magrittr in the example.
Note that although readxl is office of tidyverse, you'll even so need to load it explicitly as it's not a "core" tidyverse package.
Writing multiple Excel files
Let united states showtime off with the iris dataset that is pre-loaded with R. If you're not ane of us sorry people who about know this dataset by center, here's what it looks similar:
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species ## 1 five.i iii.v 1.4 0.2 setosa ## 2 iv.nine 3.0 1.4 0.2 setosa ## 3 4.7 3.2 1.iii 0.2 setosa ## 4 4.6 iii.1 1.v 0.two setosa ## 5 5.0 3.6 1.four 0.2 setosa ## vi 5.iv 3.9 1.7 0.4 setosa The starting time matter that we want to exercise is to create multiple datasets, which nosotros can do so by splitting iris. I'll do this by running group_split() on the Species column, so that each species of iris has its own dataset. This will return a list of three data frames, one for each unique value in Species: setosa, versicolor, and virginica. I'll assign these three data frames to a list object chosen list_of_dfs:
# Dissever: one data frame per Species iris %>% dplyr:: group_split(Species) -> list_of_dfs list_of_dfs ## [[1]] ## # A tibble: 50 10 five ## Sepal.Length Sepal.Width Petal.Length Petal.Width Species ## <dbl> <dbl> <dbl> <dbl> <fct> ## 1 5.1 3.5 i.4 0.2 setosa ## ii four.9 3 1.4 0.2 setosa ## 3 four.7 three.2 one.3 0.2 setosa ## 4 4.6 3.one 1.5 0.two setosa ## v 5 3.half dozen i.iv 0.2 setosa ## 6 5.four iii.9 1.7 0.4 setosa ## 7 iv.6 3.4 1.4 0.3 setosa ## 8 5 3.4 1.v 0.2 setosa ## nine four.4 2.9 1.four 0.2 setosa ## 10 iv.9 3.1 1.v 0.1 setosa ## # ... with twoscore more rows ## ## [[2]] ## # A tibble: 50 x 5 ## Sepal.Length Sepal.Width Petal.Length Petal.Width Species ## <dbl> <dbl> <dbl> <dbl> <fct> ## 1 7 3.2 four.vii one.4 versicolor ## ii 6.iv 3.2 4.5 1.v versicolor ## 3 half dozen.ix 3.i 4.9 1.five versicolor ## 4 5.5 2.3 4 i.3 versicolor ## 5 6.5 2.8 four.6 1.5 versicolor ## 6 5.7 2.viii 4.five 1.iii versicolor ## 7 6.3 3.iii four.7 ane.half-dozen versicolor ## viii 4.9 ii.4 3.3 ane versicolor ## 9 six.6 ii.ix iv.half dozen 1.3 versicolor ## 10 5.2 2.7 three.9 i.4 versicolor ## # ... with 40 more rows ## ## [[3]] ## # A tibble: l 10 v ## Sepal.Length Sepal.Width Petal.Length Petal.Width Species ## <dbl> <dbl> <dbl> <dbl> <fct> ## 1 6.3 3.3 half-dozen 2.v virginica ## 2 5.viii ii.vii 5.1 1.9 virginica ## 3 vii.one 3 5.9 2.one virginica ## iv half dozen.3 2.9 5.6 1.eight virginica ## v half-dozen.v iii 5.eight two.2 virginica ## half-dozen 7.6 iii six.6 2.1 virginica ## 7 4.nine two.5 4.5 i.7 virginica ## 8 7.3 2.9 6.3 ane.8 virginica ## ix vi.7 2.5 5.8 1.8 virginica ## 10 seven.ii 3.6 6.one 2.5 virginica ## # ... with xl more rows I'll likewise utilise purrr::map() to take the character values (setosa, versicolor, and virginica) from the Species cavalcade itself for assigning names to the list. map() transforms an input by applying a function to each element of the input, and then returns a vector the same length as the input. In this firsthand example, the input is the list_of_dfs and we use the function dplyr::pull() to extract the Species variable from each data frame. We and then repeat this arroyo to catechumen Species into character type with equally.character() and take out a single value with unique():
# Utilize the value from the "Species" column to provide a proper noun for the list members list_of_dfs %>% purrr:: map(~ pull(.,Species)) %>% # Pull out Species variable purrr:: map(~ every bit.grapheme(.)) %>% # Catechumen factor to character purrr:: map(~ unique(.)) -> names(list_of_dfs) # Set this as names for list members names(list_of_dfs) ## [1] "setosa" "versicolor" "virginica" These names will be useful for exporting the data frames into Excel, equally they will effectively exist our Excel canvass names. Yous can e'er manually hard-code the sheet names, but the above approach allows y'all to practice the entire affair dynamically if yous demand to.
Having ready the canvass names, I tin so pipe the list of information frames directly into write_xlsx(), where the Excel file proper noun and path is specified in the same path statement:
list_of_dfs %>% writexl:: write_xlsx(path = "../datasets/exam-excel/test-excel.xlsx") Writing multiple CSV files
Exporting the list of data frames into multiple CSV files will have a few more lines of code, but even so relatively straightforward. There are three main steps:
-
Ascertain a role that tells R what the names for each CSV file should be, which I've called
output_csv()below. The data statement will take in a data frame whilst the names argument will have in a character string that volition class part of the file proper noun for the individual CSV file. -
Create a named list where the names match the arguments of the role you've just divers (information and names), and should incorporate the objects that you would like to pass through to the function for the respective arguments. In this case, list_of_dfs volition provide the iii data frames, and names(list_of_dfs) volition provide the names of those three data frames. This is necessary for running
pmap(), which in my view is basically a super-powered version ofmap()that lets you iterate over multiple inputs simultaneously. -
pmap()will then iterate through the 2 sets of inputs throughoutput_csv()(the inputs are used as arguments), which then writes the 3 CSV files with the file names y'all desire. For the "writing" function, you could either usewrite_csv()from readr (part of tidyverse) orfwrite()from data.table, depending on your workflow / style.
# Step one # Define a function for exporting csv with the desired file names and into the right path output_csv <- function(data, names){ folder_path <- "../datasets/exam-excel/" write_csv(information, paste0(folder_path, "test-excel-", names, ".csv")) } # Step 2 list(data = list_of_dfs, names = names(list_of_dfs)) %>% # Step 3 purrr:: pmap(output_csv) The outcome of the in a higher place lawmaking is shown beneath. My directory now contains one Excel file with three Worksheets (sheet names are "setosa", "versicolor", and "virginica"), and iii split up CSV files for each data slice:
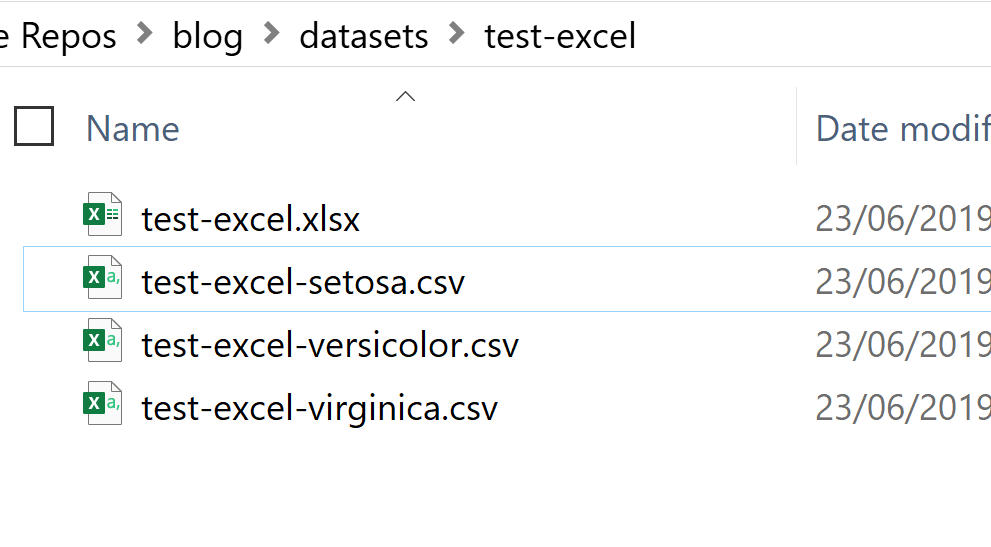
Reading multiple Excel / CSV files

For reading files in, you'll need to decide on how yous want them to exist read in. The options are:
- Read all the datasets directly into the Global Environment as individual information frames with a "divide existence" and split names.
- Read all the datasets into a single list, where each information frame is a member of that listing.
The first option is best if you are unlikely to run similar operations on all the data frames at the aforementioned fourth dimension. You may for instance want to do this if the data sets that you are reading in are structurally different from each other, and that you are planning to manipulate them separately.
The 2nd pick will be best if you are probable to dispense all the data frames at the same time, where for instance you may run on the list of data frames map() with drop_na() as an argument to remove missing values for all of the data frames at the aforementioned time. The benefit of reading your multiple data sets into a list is that you will have a much cleaner workspace (Global Environs). All the same, there is a minor and almost negligible inconvenience accessing individual data frames, as you will need to get into a listing and choice out the right member of the listing (due east.grand. doing something like list_of_dfs[3]).
Method 1A: Read all sheets in Excel into Global Surroundings
And so permit's begin! This method will read all the sheets within a specified Excel file and load them into the Global Environment, using variable names of your own choice. For simplicity, I will apply the original Excel sail names equally the variable names.
The first thing to exercise is to specify the file path to the Excel file:
wb_source <- "../datasets/test-excel/test-excel.xlsx" You tin can then run readxl::excel_sheets() to excerpt the sheet names in that Excel file, and salvage it equally a graphic symbol type vector.
# Extract the canvass names equally a graphic symbol string vector wb_sheets <- readxl:: excel_sheets(wb_source) print(wb_sheets) ## [1] "setosa" "versicolor" "virginica" The side by side step is to iterate through the sail names (saved in wb_sheets) using map(), and within each iteration use assign() (base) and read_xlsx() (from readxl) to load each individual sheet into the Global Surroundings, giving each ane a variable proper name. Here's the lawmaking:
# Load everything into the Global Surroundings wb_sheets %>% purrr:: map(function(sheet){ # iterate through each canvas proper noun assign(x = sheet, value = readxl:: read_xlsx(path = wb_source, canvass = sail), envir = .GlobalEnv) }) This is what my work space looks similar: 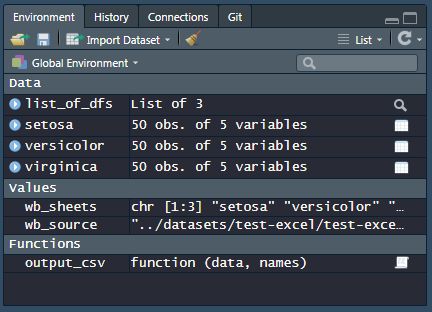
Notation that map() always returns a list, simply in this example we practice not need a list returned and only require the "side effects", i.e. the objects being read in to be assigned to the Global Surroundings. If you prefer you can use lapply() instead of map(), which for this purpose doesn't make a large practical difference.
As well, assign() allows you to assign a value to a name in an surround, where we've specified the following as arguments:
- x:
canvasas the variable proper name - value: The actual data from the sheet we read in. Hither, we use
readxl::read_xlsx()for reading in specific sheets from the Excel file, where yous simply specify the file path and the sheet name as the arguments. - envir:
.GlobalEnvequally the environment
Method 1B: Read all CSV files in directory into Global Environment
The method for reading CSV files into a directory is slightly different, equally you'll need to find a way to identify or create a character vector of names of all the files that you want to load into R. To practise this, we'll use list.files(), which produces a character vector of the names of files or directories in the named directory:
file_path <- "../datasets/test-excel/" file_path %>% listing.files() ## [1] "examination-excel-setosa.csv" "exam-excel-versicolor.csv" ## [three] "exam-excel-virginica.csv" "test-excel.xlsx" We merely want CSV files in this case, and then we'll want to practice a fleck of string manipulation (using str_detect() from stringr - again, from tidyverse) to get just the names that terminate with the extension ".csv". Let'southward pipe this along:
file_path %>% list.files() %>% .[str_detect(., ".csv")] -> csv_file_names csv_file_names ## [1] "test-excel-setosa.csv" "examination-excel-versicolor.csv" ## [three] "test-excel-virginica.csv" The side by side part is like to what we've washed earlier, using map(). Note that apart from replacing the value statement with read_csv() (or you tin can utilise fread() to render a data.tabular array object rather than a tibble), I too removed the file extension in the x argument so that the variable names would non contain the actual characters ".csv":
# Load everything into the Global Environment csv_file_names %>% purrr:: map(role(file_name){ # iterate through each file proper name assign(x = str_remove(file_name, ".csv"), # Remove file extension ".csv" value = read_csv(paste0(file_path, file_name)), envir = .GlobalEnv) }) Method 2A: Read all sheets in Excel into a list
Reading sheets into a listing is really easier than to read it into the Global Environs, as map() returns a listing and you won't have to utilize assign() or specify a variable proper name. Recall that wb_source holds the path of the Excel file, and wb_sheets is a character vector of all the sail names in the Excel file:
# Load everything into the Global Surround wb_sheets %>% purrr:: map(role(canvas){ # iterate through each canvass name readxl:: read_xlsx(path = wb_source, sheet = sail) }) -> df_list_read # Assign to a list Y'all can and then utilize map() again to run operations across all members of the list, and fifty-fifty chain operations inside it:
df_list_read %>% map(~ select(., Petal.Length, Species) %>% head()) ## [[1]] ## # A tibble: 6 x 2 ## Petal.Length Species ## <dbl> <chr> ## 1 one.4 setosa ## 2 1.4 setosa ## 3 1.3 setosa ## 4 1.5 setosa ## v ane.4 setosa ## 6 1.7 setosa ## ## [[2]] ## # A tibble: 6 ten two ## Petal.Length Species ## <dbl> <chr> ## 1 4.seven versicolor ## 2 4.five versicolor ## 3 4.9 versicolor ## iv iv versicolor ## 5 four.6 versicolor ## 6 4.5 versicolor ## ## [[three]] ## # A tibble: half dozen x ii ## Petal.Length Species ## <dbl> <chr> ## 1 6 virginica ## 2 5.1 virginica ## 3 five.9 virginica ## 4 v.six virginica ## 5 five.8 virginica ## 6 half-dozen.vi virginica Method 2B: Read all CSV files in directory into a list
At this point you've probably gathered how you can conform the lawmaking to read CSV files into a listing, but let'southward cover this for comprehensiveness. No assign() needed, and only run read_csv() within the map() function, iterating through the file names:
# Load everything into the Global Surroundings csv_file_names %>% purrr:: map(part(file_name){ # iterate through each file proper name read_csv(paste0(file_path, file_name)) }) -> df_list_read2 # Assign to a list Give thanks you for reading! 😄
Hopefully this is a helpful tutorial for an iterative arroyo to writing and reading Excel files. If you similar what yous read or if you accept whatsoever suggestions / thoughts most the subject, exercise leave a comment in the Disqus fields in the blog and permit me know!
Source: https://martinctc.github.io/blog/vignette-write-and-read-multiple-excel-files-with-purrr/
0 Response to "How to Read Multiple Excel Files in R"
Enregistrer un commentaire